Agglomerative Clustering Instance — Step by Step
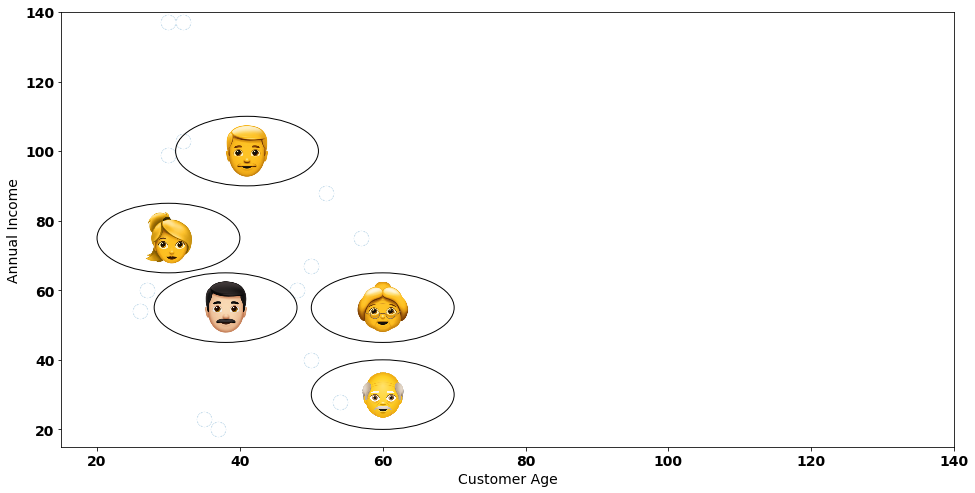
In our step-by-step instance, we’re going to use a fictional dataset with 5 clients:
Let’s think about that we run a store with 5 clients and we needed to group these clients based mostly on their similarities. We have now two variables that we wish to take into account: the buyer’s age and their annual revenue.
Step one of our agglomerative clustering consists of making pairwise distances between all our knowledge factors. Let’s do exactly that by representing every knowledge level by their coordinate in a [x, y] format:
- Distance between [60, 30] and [60, 55]: 25.0
- Distance between [60, 30] and [30, 75]: 54.08
- Distance between [60, 30] and [41, 100]: 72.53
- Distance between [60, 30] and [38, 55]: 33.30
- Distance between [60, 55] and [30, 75]: 36.06
- Distance between [60, 55] and [41, 100]: 48.85
- Distance between [60, 55] and [38, 55]: 22.0
- Distance between [30, 75] and [41, 100]: 27.31
- Distance between [30, 75] and [38, 55]: 21.54
- Distance between [41, 100] and [38, 55]: 45.10
Though we will use any kind of distance metric we would like, we’ll use euclidean resulting from its simplicity. From the pairwise distances we’ve calculated above, which distance is the smallest one?
The space between center aged clients that make lower than 90k {dollars} a yr — clients on coordinates [30, 75] and [38, 55]!
Reviewing the system for euclidean distance between two arbitrary factors p1 and p2:
Let’s visualize our smallest distance on the 2-D plot by connecting the 2 clients which might be nearer:
The following step of hierarchical clustering is to contemplate these two clients as our first cluster!
Subsequent, we’re going to calculate the distances between the info factors, once more. However this time, the 2 clients that we’ve grouped right into a single cluster might be handled as a single knowledge level. For example, take into account the crimson level beneath that positions itself in the midst of the 2 knowledge factors:
In abstract, for the subsequent iterations of our hierarchical answer, we gained’t take into account the coordinates of the unique knowledge factors (emojis) however the crimson level (the common between these knowledge factors). That is the usual method to calculate distances on the common linkage technique.
Different strategies we will use to calculate distances based mostly on aggregated knowledge factors are:
- Most (or full linkage): considers the farthest knowledge level within the cluster associated to the purpose we try to mixture.
- Minimal (or single linkage): considers the closest knowledge level within the cluster associated to the purpose we try to mixture.
- Ward (or ward linkage): minimizes the variance within the clusters with the subsequent aggregation.
Let me do a small break on the step-by-step clarification to delve a bit deeper into the linkage strategies as they’re essential in this sort of clustering. Right here’s a visible instance of the completely different linkage strategies obtainable in hierarchical clustering, for a fictional instance of three clusters to merge:
Within the sklearn implementation, we’ll have the ability to experiment with a few of these linkage strategies and see a big distinction within the clustering outcomes.
Returning to our instance, let’s now generate the distances between all our new knowledge factors — keep in mind that there are two clusters which might be being handled as a single one any longer:
- Distance between [60, 30] and [60, 55]: 25.0
- Distance between [60, 30] and [34, 65]: 43.60
- Distance between [60, 30] and [41, 100]: 72.53
- Distance between [60, 55] and [34, 65]: 27.85
- Distance between [60, 55] and [41, 100]: 48.85
- Distance between [34, 65] and [41, 100]: 35.69
Which distance has the shortest path? It’s the trail between knowledge factors on coordinates [60, 30] and [60, 55]:
The following step is, naturally, to affix these two clients right into a single cluster:
With this new panorama of clusters, we calculate pairwise distances once more! Keep in mind that we’re all the time contemplating the common between knowledge factors (as a result of linkage technique we selected) in every cluster as reference level for the space calculation:
- Distance between [60, 42.5] and [34, 65]: 34.38
- Distance between [60, 42.5] and [41, 100]: 60.56
- Distance between [34, 65] and [41, 100]: 35.69
Apparently, the subsequent knowledge factors to mixture are the 2 clusters as they lie on coordinates [60, 42.5] and [34, 65]:
Lastly, we end the algorithm by aggregating all knowledge factors in a single massive cluster:
With this in thoughts, the place will we precisely cease? It’s in all probability not a fantastic thought to have a single massive cluster with all knowledge factors, proper?
To know the place we cease, there’s some heuristic guidelines we will use. However first, we have to get conversant in one other approach of visualizing the method we’ve simply performed — the dendrogram:
On the y-axis, we have now the distances that we’ve simply calculated. On the x-axis, we have now every knowledge level. Climbing from every knowledge level, we attain an horizontal line — the y-axis worth of this line states the full distance that can join the info factors on the perimeters.
Keep in mind the primary clients we’ve linked in a single cluster? What we’ve seen within the 2D plot matches the dendrogram as these are precisely the primary clients linked utilizing an horizontal line (climbing the dendrogram from beneath):
The horizontal strains signify the merging course of we’ve simply performed! Naturally, the dendrogram ends in an enormous horizontal line that connects all knowledge factors.
As we simply received conversant in the Dendrogram, we’re now able to examine the sklearn implementation and use an actual dataset to know how we will choose the suitable variety of clusters based mostly on this cool clustering technique!
Sklearn Implementation
For the sklearn implementation, I’m going to make use of the Wine High quality dataset obtainable right here.
wine_data = pd.read_csv('winequality-red.csv', sep=';')
wine_data.head(10)
This dataset incorporates details about wines (notably crimson wines) with completely different traits reminiscent of citric acid, chlorides or density. The final column of the dataset offers respect to the standard of the wine, a classification performed by a jury panel.
As hierarchical clustering offers with distances and we’re going to make use of the euclidean distance, we have to standardize our knowledge. We’ll begin by utilizing a StandardScaleron prime of our knowledge:
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
wine_data_scaled = sc.fit_transform(wine_data)
With our scaled dataset, we will match our first hierarchical clustering answer! We are able to entry hierarchical clustering by creating an AgglomerativeClustering object:
average_method = AgglomerativeClustering(n_clusters = None,
distance_threshold = 0,
linkage = 'common')
average_method.match(wine_data_scaled)
Let me element the arguments we’re utilizing contained in the AgglomerativeClustering:
n_clusters=Noneis used as a method to have the complete answer of the clusters (and the place we will produce the complete dendrogram).distance_threshold = 0should be set within thesklearnimplementation for the complete dendrogram to be produced.linkage = ‘common’is a vital hyperparameter. Keep in mind that, within the theoretical implementation, we’ve described one technique to contemplate the distances between newly shaped clusters.commonis the tactic that considers the common level between each new shaped cluster within the calculation of recent distances. Within thesklearnimplementation, we have now three different strategies that we additionally described:single,fullandward.
After becoming the mannequin, it’s time to plot our dendrogram. For this, I’m going to make use of the helper perform offered within the sklearn documentation:
from scipy.cluster.hierarchy import dendrogramdef plot_dendrogram(mannequin, **kwargs):
# Create linkage matrix after which plot the dendrogram
# create the counts of samples underneath every node
counts = np.zeros(mannequin.children_.form[0])
n_samples = len(mannequin.labels_)
for i, merge in enumerate(mannequin.children_):
current_count = 0
for child_idx in merge:
if child_idx < n_samples:
current_count += 1 # leaf node
else:
current_count += counts[child_idx - n_samples]
counts[i] = current_count
linkage_matrix = np.column_stack(
[model.children_, model.distances_, counts]
).astype(float)
# Plot the corresponding dendrogram
dendrogram(linkage_matrix, **kwargs)
If we plot our hierarchical clustering answer:
plot_dendrogram(average_method, truncate_mode="stage", p=20)
plt.title('Dendrogram of Hierarchical Clustering - Common Methodology')
The dendrogram shouldn’t be nice as our observations appear to get a bit jammed. Generally, the common , single and full linkage could lead to unusual dendrograms, notably when there are sturdy outliers within the knowledge. The ward technique could also be applicable for this sort of knowledge so let’s check that technique:
ward_method = AgglomerativeClustering(n_clusters = None,
distance_threshold = 0,
linkage = 'ward')
ward_method.match(wine_data_scaled)plot_dendrogram(ward_method, truncate_mode="stage", p=20)
Significantly better! Discover that the clusters appear to be higher outlined in line with the dendrogram. The ward technique makes an attempt to divide clusters by minimizing the intra-variance between newly shaped clusters (https://on-line.stat.psu.edu/stat505/lesson/14/14.7) as we’ve described on the primary a part of the put up. The target is that for each iteration the clusters to be aggregated decrease the variance (distance between knowledge factors and new cluster to be shaped).
Once more, altering strategies might be achieved by altering the linkage parameter within the AgglomerativeClustering perform!
As we’re proud of the look of the ward technique dendrogram, we’ll use that answer for our cluster profilling:
Are you able to guess what number of clusters we must always select?
In response to the distances, a very good candidate is chopping the dendrogram on this level, the place each cluster appears to be comparatively removed from one another:
The variety of vertical strains that our line crosses are the variety of last clusters of our answer. Selecting the variety of clusters shouldn’t be very “scientific” and completely different variety of clustering options could also be achieved, relying on enterprise interpretation. For instance, in our case, chopping off our dendrogram a bit above and lowering the variety of clusters of the ultimate answer might also be an speculation.
We’ll stick to the 7 cluster answer, so let’s match our ward technique with these n_clusters in thoughts:
ward_method_solution = AgglomerativeClustering(n_clusters = 7,
linkage = 'ward')
wine_data['cluster'] = ward_method_solution.fit_predict(wine_data_scaled)
As we wish to interpret our clusters based mostly on the unique variables, we’ll use the predict technique on the scaled knowledge (the distances are based mostly on the scaled dataset) however add the cluster to the unique dataset.
Let’s examine our clusters utilizing the means of every variable conditioned on the cluster variable:
wine_data.groupby([‘cluster’]).imply()
Apparently, we will begin to have some insights in regards to the knowledge — for instance:
- Low high quality wines appear to have a big worth of
whole sulfur dioxide— discover the distinction between the best common high quality cluster and the decrease high quality cluster:
And if we examine the high quality of the wines in these clusters:
Clearly, on common, Cluster 2 incorporates larger high quality wines.
One other cool evaluation we will do is performing a correlation matrix between clustered knowledge means:
This offers us some good hints of potential issues we will discover (even for supervised studying). For instance, on a multidimensional stage, wines with larger sulphates and chlorides could get bundled collectively. One other conclusion is that wines with larger alcohol proof are typically related to larger high quality wines.